
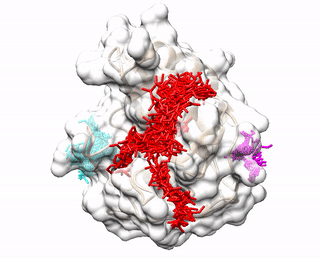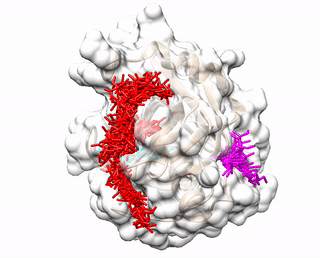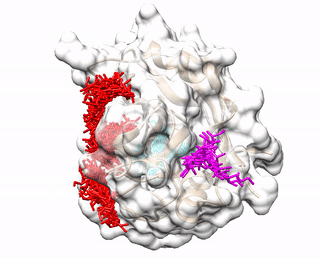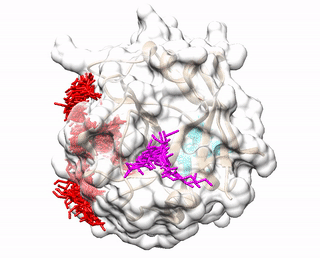
ACCLUSTER predicts peptide binding sites on a given protein structure by clustering chemical interactions between amino acid probes and the protein receptor.
Please go to the "About" page for the details about the ACCLUSTER.
To submit a job, at least one input, a ".pdb" file of the protein structure, is required. Other inputs are optional, such as the sequence of the bound peptide and
a ."txt" file that specifies the residue(s) remote from the binding site to be blocked in the computation.
Detailed instructions about job submission and prediction results are described on the "Tutorial" page.
Please cite the following references for ACCLUSTER:
C. Yan, X. Xu, X. Zou. The Usage of ACCLUSTER for Peptide Binding Site Prediction. Modeling Peptide-Protein Interactions: Methods and Protocols, 3-9, 2017.
C. Yan, X. Zou. Predicting peptide binding sites on protein surfaces by clustering chemical interactions. J. Comput. Chem, 36: 49-61, 2015. (featured in the inside cover of the issue).