
1. Required inputs
1.1 Protein receptor structure
The server requires only the PDB file of the protein structure (one chain or multiple chains) as the input. Although
the uploaded protein file will be cleaned (e.g. deletion of the solvates, ligands and hydrogen atoms) after submission, uploading a
pre-checked structure is recommended. The protein should contain 31-1000 amino acids.
2. Advanced options (optional)
2.1 Peptide sequence
The users are allowed to provide the sequence of the bound peptide. The peptide sequence can be provided either by entering single-letter
amino acid codes or by uploading a FASTA file. The sequence length of the protein should be within 4-30.
2.2 Residues to be blocked:
ACCLUSTER allows users to block residue(s) that are known to be remote from the binding site. These residues are specified in a ".txt"
file that copies the columns 18 to 26 of the corresponding atom lines in the protein pdb file, as shown in the following example.
3. An example of the inputs
(PDB: 1AWQ):
Protein receptor structure: 1AWQ_rec.pdb
Peptide sequence (FASTA file): 1AWQ_pep.fasta(optional)
Blocked residues (FASTA file): blocked.txt(optional)
4. Job status
After a job is submitted, the ACCLUSTER server will immediately examine the uploaded files. A pop-up page will inform the user
whether the job is successfully submitted. If not, the ERROR information will be reported on this page.
Once the job is successfully submitted, the job status is monitored on the "Queue" page.
When the job is waiting for computational resource allocation, the job status is "Pending".
Once the job starts to run, the status
will be changed to "Running".
The user will receive an email notification after the job is completed, if the email address is provided. Then, the status will be changed to "Done" or "Error".
Note: If the user selects the option "Do not show my job on the queue page" on the submission page, the job status
will not appear on the "Queue" page. In this case, the only way to know the status is through email notification.
4. Results
4.1 How to receive the results
When the job is completed, a result page will be created. There are three ways to obtain the link of the result page:
a) The link is provided on the pop-up page after the job is successfully submitted.
b) The link will be sent to the user by email when the job is completed.
c) The user can find the job on the "Queue" page through keyword search, and then click the corresponding "Job ID".
4.2 The result page
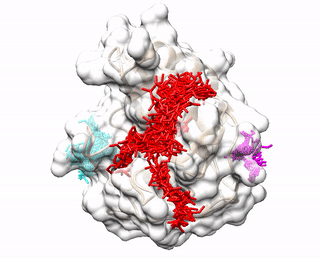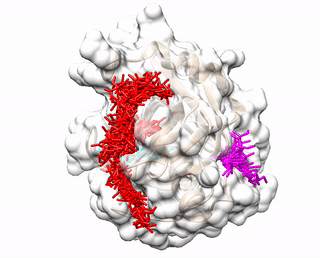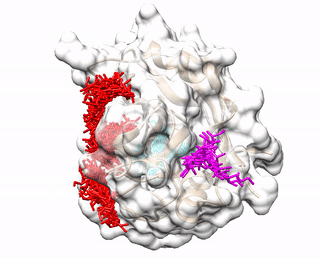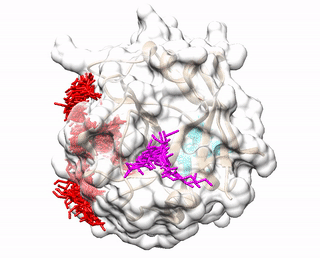
Up to three predicted binding sites are provided to users. These binding sites and the receptor are displayed via a JavaScript library 3Dmol.js. The top prediction is colored red. The second prediction is colored cyan. The third prediction is colored magenta. The pdb files (accluster_site1.pdb, accluster_site2.pdb, accluster_site3.pdb ) representing the predicted binding sites can be downloaded, and visualized using molecular visualization programs such as Chimera.