
About MDockPeP2
The MDockPeP2 server predicts protein-peptide complex structures starting with the protein structure and the peptide sequence.
A flowchart of the protocol is shown as following.
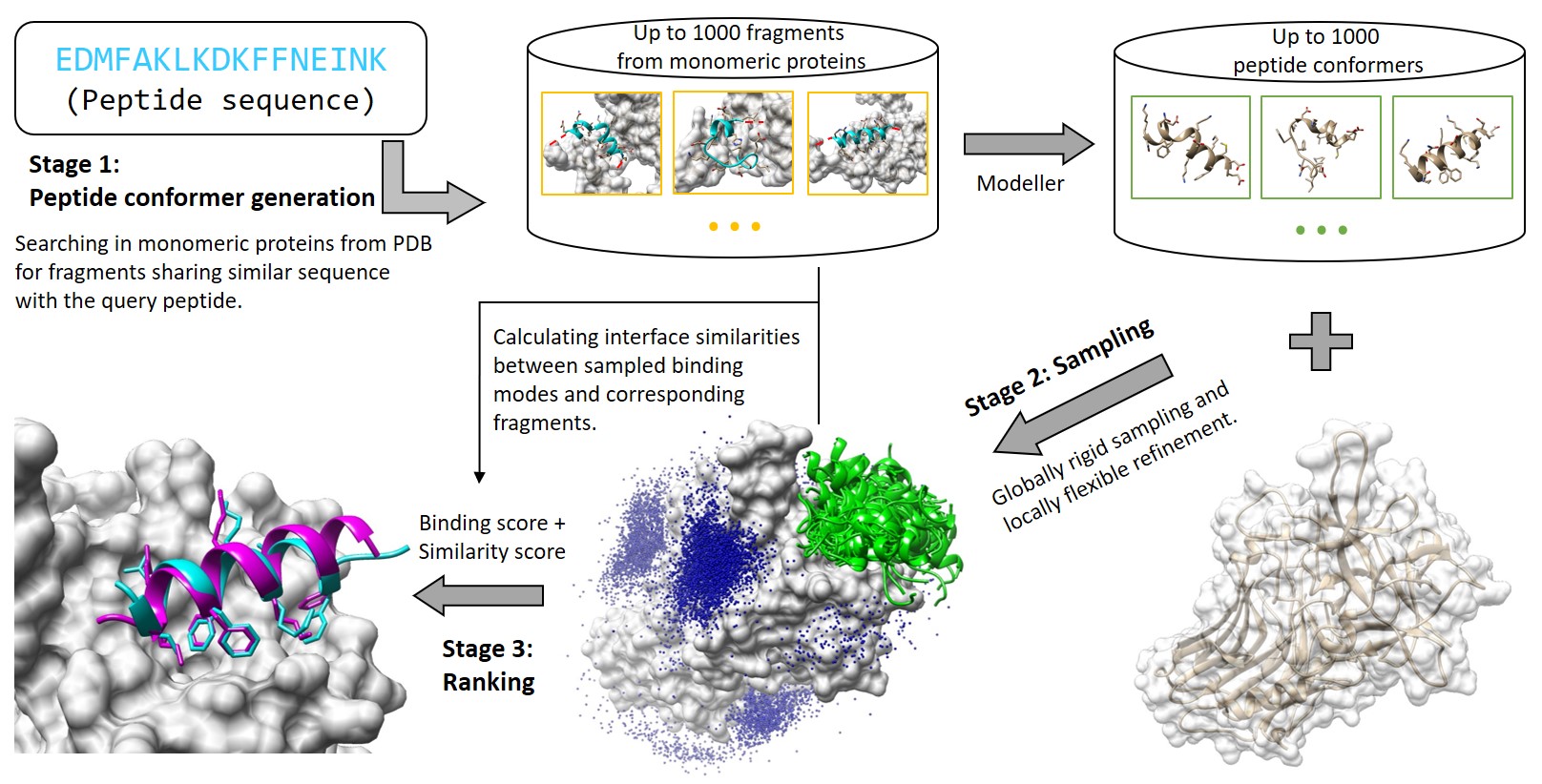
MDockPeP2 can be briefly divided into three stages.
-
In Stage 1, for a query peptide, a protein structure database is searched to find fragments sharing similar sequence. These fragments are used as templates to build peptide conformers.
-
In Stage 2, peptide conformers are generated and then independently docked to a target protein surface using a well-developed protein-protein docking program (global rigid docking). The generated models are further refined using a well-established protein-small molecule docking program (local flexible refinement).
-
In Stage 3, peptide conformers are merged and evaluated using a hybrid scoring function, which considers both calculated binding scores and interface similarities.
More details about MDockPeP2 are available in the reference:
- Xu X, Zou X. Predicting Protein-peptide complex structures by accounting for peptide flexibility and physicochemical environment. J. Chem. Inf. Model., 2022, 62: 27-39. DOI: https://doi.org/10.1021/acs.jcim.1c00836